Digital in-line holography is a 3D microscopy technique which has gotten an increasing amount of attention over the last few years in the fields of microbiology, medicine and physics. The fully-automatic analysis of such microscopic data is highly challenging, especially since the motion patterns of observed microorganisms can change rapidly over time.
The project is divided in two parts: tracking and classification.For the first part of the project, we approach the challenges of a high throughput analysis of holographic microscopy data and present a system for detecting particles in 3D reconstructed holograms and their 3D trajectory estimation over time. Our main contribution is a robust method which evolves from the Hungarian bipartite weighted graph matching algorithm and allows us to deal with newly entering and leaving particles and compensate for missing data and outliers. In the experiments we compare our fully automatic system with manually labeled ground truth data and we can report an accuracy between 76% and 91%.
For the second part of the project, we deal with the classification of complex microorganism motion patterns observed with this microscopy technique. We use of Hidden Markov Models (HMM) to classify four different motion patterns of a microorganism and to separate multiple patterns occurring within a trajectory. We perform leave-one-out experiments with the training data to prove the accuracy of our method and to analyze the importance of each trajectory feature for classification. We further present results obtained on four full sequences, a total of 2500 frames. The classification rates obtained range between 83.5% and 100%.
An alternative to conventional optical microscopy is provided by digital in-line holography, a lensless microscopy technique which intrinsically contains three-dimensional information about the volume under investigation.
The important requirements are a sample of sufficient transparency and a divergent, coherent wave. The holographic microscope setup follows directly Gabors initial idea and has been implemented for laser radiation by Xu et al.
A wavefront is generated by diffraction of a laser beam at a pinhole. A so-called hologram is then recorded by a CCD- or CMOS-chip.
This hologram is then reconstructed back into real space by a Kirchhoff-Helmholtz transformation.
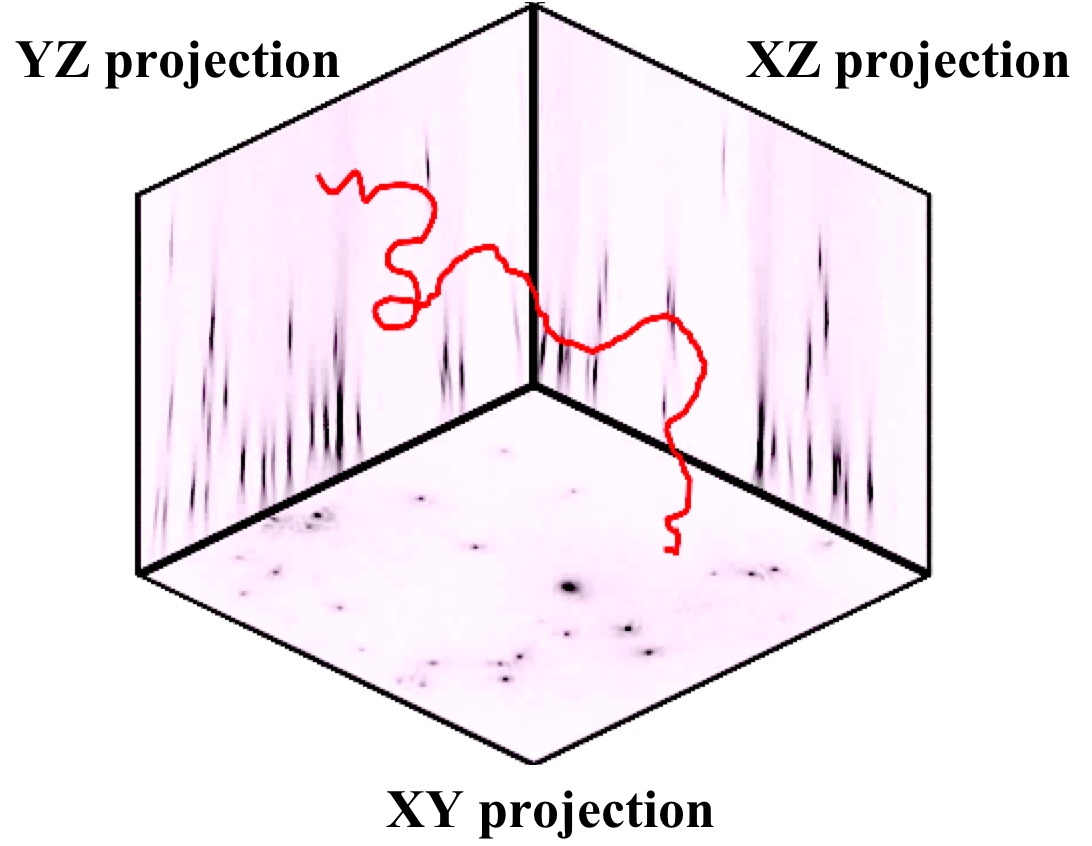
From the reconstruction 3 projections XY, XZ and YZ are obtained (see Figure).
These projections contain the image information of the complete observation volume, i.e. from every object located in the light cone between pinhole and detector.
As any tracking algorithm, we first need to detect the position of the particles on every frame. In order to deal with the noise present in the images, we apply a Laplacian of Gaussian (LoG) filter which will bring out the "round" microorganisms we want to detect. Of course, this filter can be adapted according to the shape of the particle we need to detect. We then use thresholding to detect the particles.
In order to match the particles from one frame to the next and recover the full trajectory, we make use of the Hungarian matching algorithm. With this method, we encounter 3 important problems:
The cost function as key input for the Hungarian algorithm is created using the Euclidean distances between particles. The first problem is easily solved by using a threshold on the cost matrix. This threshold depends on the observed maximum speed of the microorganism under study.
For the second problem, we make use of IN/OUT states to model the possible entrances and exits of the particles from the Field Of View (FOV) of the holographic microscope.
The third problem is the most difficult to handle and it can generate ID switches between the particles. To deal with it, we create a robust method which we call the Multi-level Hungarian, which consists in applying the Hungarian bipartite matching algorithm in several levels as shown in the Figure below.
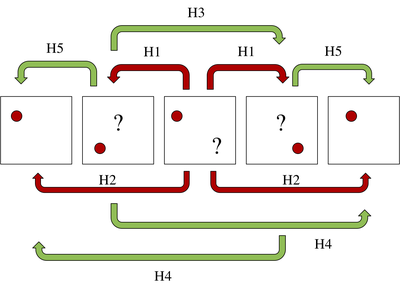
Our algorithm is able to deal with missing data and false detections. The overall method detects between 76\% and 91\% of the particles present in the volume.
Once the trajectories have been correctly detected, we perform classification into 4 different motion patterns. This way, we can automatically obtain useful information about the behavior of certain microorganisms under different environment conditions.
To perform the classification, we make use of Hidden Markov Models (HMMs) and we analyze several features to determine which are the best suited for classification.
We determine that speed, the angle between velocities and the distance to the surface are the most relevant features to successfully identify the 4 motion patterns (orientation, wobbling, gyration and spinning). For more information on the types of patterns and experimental setup, please visit the webpage of our collaborators.
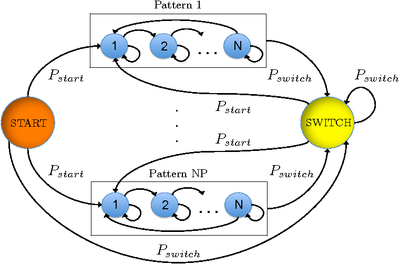
Once we have one HMM trained for each motion pattern, we build a complete HMM with an added SWITCH and a START state. With the complete HMM we can not only classify the sequences where there is one pattern present, but also sequences where the particle makes transitions between different patterns. The complete HMM is depicted in the Figure.
Experiments performed on several sequences show that our complete HMM can classify between 83.5\% and 100\% of the trajectories. We are also able to perfectly determine the moment where the transitions occur.
This work is partially funded by the German Research Foundation, DFG projects RO 2497/7-1 and RO 2524/2-1 and the EU project AMBIO.