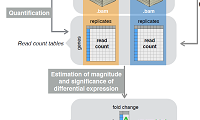
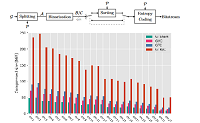
In recent years, technological advances in DNA sequencing have led to faster and more cost-efficient approaches for sequencing individual genomes and other genomic samples. Due to the enormous amount of sequencing data generated, the processing, storage, and analysis of sequencing data presents new challenges to the scientific community. New methods and tools need to be developed to improve understanding of the underlying biology and overcome current limitations in terms of storage space, processing speed, and many more.